pIII-Defective Helper Phage: M13KO7d3 & CM13d3
Helper phage designed for multivalency and improved panning efficiency.
Phage display systems of type 3+3 contain two sources of protein pIII, a recombinant fusion protein made by the phagemid and a wild-type protein III made by the helper (1). The resulting phage particles have a mosaic of proteins III, of which only a fraction are fused with antibodies. Sometimes display is very low with no more than 1% or less of all pIII bearing antibodies, making the display essentially monovalent. The use of a helper where the protein pIII coding sequence has been deleted forces the system to use the phagemid as the only source of protein pIII, resulting in an increase in the absolute number of antibodies per phage particle (2-4). The augmented avidity translates into better panning efficiency, allowing efficient isolation of positive clones where classical systems typically fail.
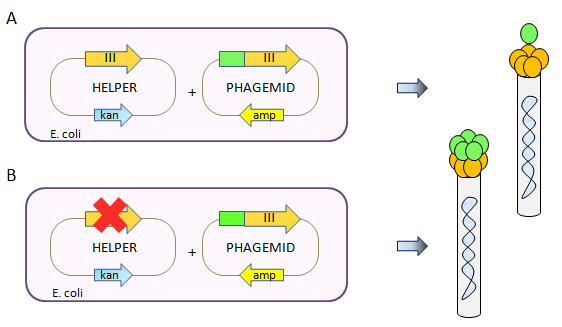
Figure 1. Defective Helper Phage System. A type 3+3 phage display system has 2 sources of protein pIII resulting in mosaic virions displaying the fusion protein in a low-valency configuration (Top panel A). The use of a pIII-defective helper phage forces the system to use the phagemid as the only source in protein pIII, resulting in a high valency display on the virion progeny (Bottom panel B).
Properties of pIII-defective helper phages
-
Improved display.
-
More efficiency panning.
-
Selection of lower affinity clones.
-
Infectivity linked to display.
-
Induction of polyphages.
Improved display
Only pIII fusion bearing antibodies are recruted during virion assembly resulting in more display of antibodies; enhancement up to 100-fold and more have been reported (4).
More efficient panning
As a consequence of better display, panning is more efficient, resulting in a higher percentage of positive clones appearing earlier in the screen.
Selection of lower affinity clones
Increase in pIII display leads to multivalent display since virions can bear up to five copies of protein pIII; the augmented avidity improves binding of lower affinity clones.
Infectivity linked to display
Because the phagemid is the only source of protein pIII and pIII is involved in bacterial transduction, the occurence of a successful transduction requires a recombinant protein pIII. Systems linking display and infectivity are known to give a lower background and faster enrichments during panning.
Induction of polyphages
pIII-defective helper phage have a tendency to induce the formation of polyphages. In a situation where display is difficult to achieve, polyphages, i.e. virions containing more than one unit of phage ssDNA, can be favored. Analysis of virions produced with a pIII-defective helper phage by Western blot often have less protein III compared to a wild-type control; the “missing” pIII most likely results from blotting polyphages with a low content in protein pIII.
CM13d3 defective Helper
The design of CM13d3 follows VCSM13d3 (3) and hyperphage (4) with an almost complete deletion of protein pIII ORF but retention of the pIII promoter and the leader peptide sequence to limit polar effects; the sequence in GenBank format is available here. The natural pVI RBS is conserved like with hyperphage. Exogenous protein pIII to enable proper packaging is provided by an accessory low copy number plasmid that does not package. Phage production is high both as a single phage or as a helper, usually making around 50% of PEG-precipitable material normally obtained with the parent helper CM13. In that regard, CM13d3 much behaves like hyperphage (6).
M13Ko7d3 defective Helper
As of April 2019, M13KO7d3 is being replaced by CM13d3. CM13d3 contains the mutation ir3B A->G at position 8418 of M13KO7 that is found in both VCSM13 and CM13 and confers a higher infectivity. As a result CM13d3 has a higher pfu/ml titer than M13KO7d3 and is more efficient at building libraries.
References
-
Smith, G. P., & Petrenko, V. a. (1997). Phage Display. Chemical Reviews, 97(2), 391–410.
-
Dueñas, M. AND Borrebaeck CA. (1995). Novel helper phage design: intergenic region affects the assembly of bacteriophages and the size of antibody libraries, FEMS Microbiol lett., 125(2-3):317-21.
-
Rakonjac, J., Jovanovic, G. & Model, P. (1997). Filamentous phage infection-mediated gene expression: construction and propagation of the gIII deletion mutant helper phage R408d3. Gene 198, 99–103 .
-
Rondot, S., Koch, J., Breitling, F. and Dübel, S. (2001). A helper phage to improve single-chain antibody presentation in phage display. Nat Biotechnol, 19(1):75-8.
-
Broders O, Breitling F, Dübel S (2003). Hyperphage – Improving antibody presentation in phage display, Methods Mol Biol, 205, 295-302.
-
Soltes G, Hust M, Ng KKY, Bansal A, Field J, Stewart DIH, Dübel S, Cha S, Wiersma EJ (2007). On the influence of vector design on antibody phage display, J Biotechnol. 127, 626-637.
PUB008 – Rev150813
Please send comments to info@abdesignlabs.com.
Copyright © 2015 Antibody Design Labs. All rights reserved. The reuse or reproduction of any of the information, design or layout contained in this web site without the permission of Antibody Design Labs is prohibited.
