MIMTM Peptide Libraries
Premade Phage Display Peptide Libraries
T
The MIM™ phage display libraries are premade peptide libraries ready for screening. Based on the phage system developed by George Smith, the MIM™ libraries are easy to screen. They do not require plaque formation and don’t need a helper phage. With a pentavalent display, they mimic naive IgM and are capable of detecting binding in the micromolar range, making them one of the best tool to discover de novo peptide binders on any target
Overview
-
Display on the p3 minor coat protein of filamentous bacteriophage.
-
Libraries built on fADL-2blue vector, ampicillin-resistant, colony-based.
-
Libraries 1 x 109 in size.
-
Diversity with trimer codons (cyclic libraries) or NNK-based (linear libraries).
-
Easy to use, sensitive to micromolar affinities.
-
No royalties, freedom to operate.
-
A license may be required for commercial use, please contact us.
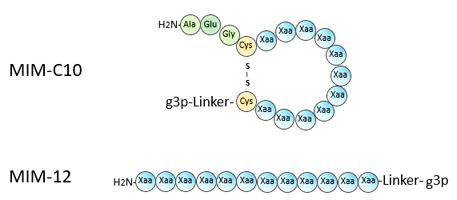
Peptide Phage Display
P
hage display was invented in 1985 by Nobel Prize winner George P. Smith in 1985 (1). This technique enables rapid in vitro molecular evolution by linking polypeptides fused to a bacteriophage coat protein with the encoding DNA found the bacteriophage genome. Using biopanning, a progressive enrichment process based on repeated rounds of selection, large collections of variants can be interrogated for specific properties. One of the first applications was the identification of antibody epitopes using a library of short random peptides (2). The method expanded rapidly to the discovery of novel binding peptides, peptide mimics such as streptavidin binders, identification of enzyme substrates, the discovery of potential therapeutic peptides, and many more applications. Due to their small size, peptides selected from phage libraries tend to bind biologically relevant sites on the surface of target proteins, making them an attracttive starting point for drug development (3).
P
hage display only requires a bench, the ability to grow E. coli in the lab, and skills in molecular biology. In such minimal settings, libraries as large as 1010 random peptides can be built and screened. Since its inception, the field has expanded with more complex library designs, introduction of cysteines to create cyclic peptides, unnatural amino acids to expand the genetic code, or post-translational modifications that alter amino acids or introduce scaffolds. Still, the method is an art full of pitfalls and traps. One of the main goals of Antibody Design Labs is to provide tools and reagents to practice this art of phage display with limited errors or downfalls.
Applications
A
pplications of phage display peptide libraries are multiple. Here are a few classical examples (reviewed in 3-6).
Drug discovery for the identification of biologically active peptides
-
Screening for receptor agonists and antagonists
-
Identification of target binding peptides
-
Anti-viral/microbial peptides
-
Identification of peptide mimics of non-peptide ligands
-
Mirror image phage display using reverse D-polypeptides
-
Agents for targeted delivery of drugs, cell-targeting, or gene delivery
Target Discovery
-
In vivo panning (vascular targeting)
-
Cell panning (novel targets)
-
Drug target validation
Proteomics
-
Mapping protein-protein interactions
Immunology
-
Epitope mapping
-
Development of vaccines, mimotopes
Enzymology
-
Substrate phage
-
Identification of enzyme substrates and inhibitors
Material
-
Material-specific peptides, e.g. ice binding peptides, etc.
-
Landscape libraries
Phage Systems for Peptide Display
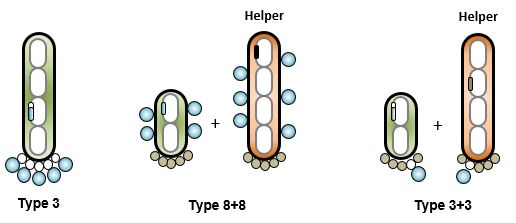
S
everal methods and systems that achieve peptide phage display have been reported. The use of phagemids in 3+3 configuration is common. Very large libraries, above 1010 are easy to build. Unfortunately, the low valency limits discovery to peptides with relatively high affinities. Some groups prefer the use of an 8+8 system to increase the valency while staying with a phagemid system (7). Type 3 systems are the preferred method for multivalent display. The downside is the large size of the vector, which makes libraries above 109 difficult to build. The very first libraries were built on derivatives of fd-tet (2). In this system, the defect in the origin of replication of the minus strand results in a colony phenotype with limited toxicity of peptide-p3 fusions (8). fADL-2blue is an ampicillin-resistant derivative of fd-tet. The MIM peptide libraries use this vector and therefore are recommended for de novo peptide discovery with affinity in the micromolar range. M13KE is a 3+3 system that is plaque-based. It has gained popularity in recent years due to the commercial availability of peptide libraries built on it. The high valency is compatible with the discovery of low affinity peptides, but the virulence of the vector and need of plaque formation enhance toxicity and limit diversity during screening campaigns.
|
Vector |
Parent |
Display |
Screening |
Description |
|---|---|---|---|---|
| fADL-2blue | Fd-tet | 3 | Colonies |
Phage with a low rfDNA copy number due to a defect in the minus origin of replication resulting in colony formation rather than plaques. High valency compatible with peptide discovery with affinities in the micromolar range. Tolerant to toxicity, preserves diversity during screening. |
| M13KE | M13mp9 | 3 | Plaques |
Virulent phage making large plaques. High valency compatible with peptide discovery with affinities in the micromolar range. Toxicity results in lower virion production and smaller to invisible plaques, limiting diversity during screening. |
| Phagemids | pUC18 | 3+3 | Colonies |
Toxicity controlled through induction. Requires helper phage. Low valency limits discovery to high affinity peptides. This system is not ideal for de novo peptide discovery. |
Library Kits
Kit Component
|
Component |
Description |
Amount |
|---|---|---|
|
MIM™ LIBRARY |
100 μl phage, ~ 2 x 1012 cfu/ml. Supplied in TBS with 10% glycerol |
100 µl |
|
PHI8S3 PRIMER |
5’-CAAGCTGTTTAAGAAATTCACCTCG, 1 nmol, 10 µM, 10 pmol/µl |
100 µl |
|
PSIR2 PRIMER |
5’-CGTTAGTAAATGAATTTTCTGTATGAGG, 1 nmol, 10 µM, 10 pmol/µl |
100 µl |
|
TG1 |
Phage-Competent™ TG1 cells, 20 OD600/ml, >2 x 1010 cfu/ml |
0.5 ml |
|
SS320 |
Phage-Competent™ SS320 cells, 20 OD600/ml, >2 x 1010 cfu/ml |
0.5 ml |
Ordering
|
Product |
Composition |
Amount |
|---|---|---|
|
MIM™-C10 Phage Display Peptide Library |
1 kit |
|
|
MIM™-12 Phage Display Peptide Library |
1 kit |
Related Products
|
Product |
Composition |
Amount |
|---|---|---|
|
fADL™-2blue Phage Vector. 20 µl at 0.5 µg/µl of DNA vector in DNA Conservation Buffer (Tris-HCL 5 mM, EDTA 0.1 mM, pH 8.5) |
10 µg |
|
|
Phage-Competent™ TG1 cells, 20 OD600/ml, >2 x 1010 cfu/ml |
0.5 ml |
|
|
Phage-Competent™ SS320 cells, 20 OD600/ml, >2 x 1010 cfu/ml |
0.5 ml |
Bulk Ordering
Please contact us for large volumes, bulk orders.
References
-
Smith, G.P. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science 1985, 228, 1315-1317.
-
Scott JK, Smith GP. Searching for peptide ligands with an epitope library. Science 1990, 249(4967):386-90.
-
Grøn H, Hyde-DeRuyscher R. Peptides as tools in drug discovery. Curr Opin Drug Discov Devel. 2000; 3(5):636-45.
-
Smith, G.P.; Petrenko, V.A. Phage Display. Chem. Rev. 1997, 97, 391-410.
-
Molek P, Strukelj B, Bratkovic T. Peptide phage display as a tool for drug discovery: targeting membrane receptors. Molecules. 2011 ;16(1):857-87.
-
Pitt A, Nims Z. Peptide Display Technologies. Methods Mol Biol. 2019; 2001:285-298.
-
Sidhu SS, Lowman HB, Cunningham BC, Wells JA. Phage display for selection of novel binding peptides. Methods Enzymol. 2000; 328:333-63.
-
Smith GP. Filamentous phage assembly: morphogenetically defective mutants that do not kill the host. Virology. 1988; 167(1):156-65.
