Custom Phage Display Peptide Library Construction
Overview
Phage display peptide libraries are a major drug discovery tool to identify de novo peptide ligands and explore sequence space in proteins. With more than 35 years of experience in the field, we are offering our expertise in the construction of peptide libraries on phage as a service for your research and development efforts. Your library will be designed with introduction of diversity precisely controlled and parameters that determine selection carefully chosen and finally build using robust methods to achieve the target size.
Vector Design & Display Type
A large choice of vectors is available to display your libraries in varied formats and types:
- High-Density Multivalent Display on the major coat protein VIII using the phagemid pADL-8.
- Multivalent Display on the minor coat protein III using the phage vector fADL-2blue. fADL-2Blue is a type 3 vector derived from fd-tet (Zacher 1980) and similar to the fUSE5 vector that was used to build the first peptide library (Scott 1990).
- Monovalent Display on the minor coat protein III using the phagemid vector pADL-100 or any other of our type 3+3 phagemids.
- Your Vector. We can accommodate you own vector to build your library.
Codon Design and Mutagenesis Strategy
We carry several methods to control the diversity introduced at each position of the sequence and can mixed them to optimize the quality of your library. A limited repertoire with a well-represented sequence space is always better than very large, degenerated libraries containing high levels of redundant codons and junk sequences source of non-specific binding.
- Hard Randomization using NNK (or NNS) codons. This is the method of choice to build brut degenerated positions or libraries that maximize the number of amino acids (all 20) but limit stop codons and redundant codons;
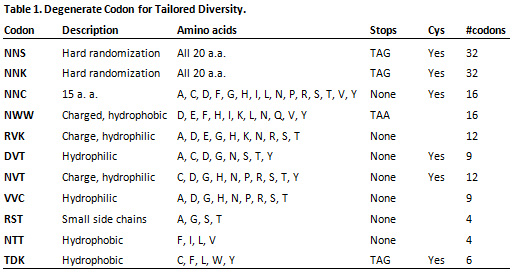
- Tailored Randomization using specialized codons encoding subsets of amino-acids with desired properties (see examples in Table 1).
- Soft Randomization by introducing a small amount of N mixture at varied positions of the wild-type sequence. This method can introduce a mutation rate high enough to generate codons with double mutations resulting in a better exploration of the sequence space than mutagenic PCR.
- Diversification to a subset of codons, usually 20 natural amino acids-encoding codons excluding stop codons and /or cysteines, using trimer codon phosphoramidites..
Library Size & Deliverables
We routinely build libraries up to 10(10) transformants depending on the number of variable positions and the degree of sequence space to be analyzed. Deliverables, including frozen cell stocks, purified DNA and purified virions, are customizable and tailored to your project. NSG analysis is available upon request.
Cost & Turnaround Time
P
lease, request a specific quote by completing the online quotation form accessible by pressing the following button; you will be contacted back by the next business day.
For all other inquiries, contact us by email at info@abdesignlabs.com or by phone at 1-877-223-3104 (PST). We guarantee competitive pricing.
All our work is performed by highly experienced staff at our San Diego facility in California. We use highly efficient cloning techniques to ensure timely delivery and have very high standards for validation. All products and services are for research use only and are not intended for use in humans.
References
- Zacher 3rd, A. N., Stock, C. A., Golden 2nd, J. W., & Smith, G. P. (1980). A new filamentous phage cloning vector: fd-tet. Gene, 9(1-2), 127–140.
- Scott JK, Smith GP. (1990). Science. 1990 Jul 27;249(4967):386-90.

